Import packages
Load sample dataset
## start stop event age year surgery transplant id
## 1 0 50 1 -17.155373 0.1232033 0 0 1
## 2 0 6 1 3.835729 0.2546201 0 0 2
## 3 0 1 0 6.297057 0.2655715 0 0 3
## 4 1 16 1 6.297057 0.2655715 0 1 3
## 5 0 36 0 -7.737166 0.4900753 0 0 4
## 6 36 39 1 -7.737166 0.4900753 0 1 4
Performing survival analysis with heart dataset
survival <- Surv(heart$start, heart$stop, heart$event == 1) # creating survival object
surv_fit <- survfit(survival ~ 1) # fit a survival curve
plot(surv_fit, main = "Kaplan Meier Plot (heart dataset)")
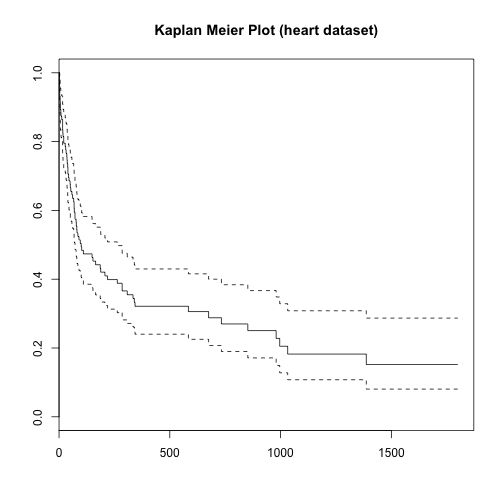
## $n
## [1] 172
##
## $time
## [1] 1 2 3 5 6 8 9 12 16 17 18 21 28 30
## [15] 32 35 36 37 39 40 43 45 50 51 53 58 61 66
## [29] 68 69 72 77 78 80 81 85 90 96 100 102 110 149
## [43] 153 165 186 188 207 219 263 285 308 334 340 343 584 675
## [57] 733 852 980 996 1032 1387
##
## $n.risk
## [1] 103 102 99 96 94 92 91 89 88 85 84 83 81 80 78 77 76
## [18] 75 74 72 70 69 68 67 66 65 64 63 62 60 59 57 56 55
## [35] 54 53 52 51 50 49 47 45 44 43 41 40 39 38 37 35 33
## [52] 32 31 29 21 17 16 14 11 10 9 6
##
## $n.event
## [1] 1 3 3 2 2 1 1 1 3 1 1 2 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 2 1 2 1 1 1 1
## [36] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1
##
## $n.censor
## [1] 2 3 3 5 1 2 0 6 2 2 1 5 8 0 5 1 2 2 2 0 2 0 2 2 0 2 1 0 2 0 1 0 1 0 0
## [36] 2 0 1 0 0 1 2 0 1 1 0 0 1 0 1 0 1 1 0 7 3 0 1 2 0 0 2
##
## $n.enter
## [1] 105 3 3 5 1 2 0 5 2 2 1 5 8 0 4 1 2
## [18] 2 1 0 2 0 2 2 0 2 1 0 2 0 1 0 1 0
## [35] 0 2 0 1 0 0 0 1 0 1 0 0 0 1 0 0 0
## [52] 1 0 0 0 0 0 0 0 0 0 0
Perform survival analysis against heart surgery status
heart$surgery <- as.factor(heart$surgery)
surv_fit2 <- survfit(survival ~ heart$surgery) # fit a survival curve
plot(surv_fit2, main="Kaplan Meier Plot by Surgery", xlab="Time in Days", ylab="Survival Proportion", col=c(1:2))
legend('topright', c("Surgery: No","Surgery: Yes"), lty=1, col=c(1:2))
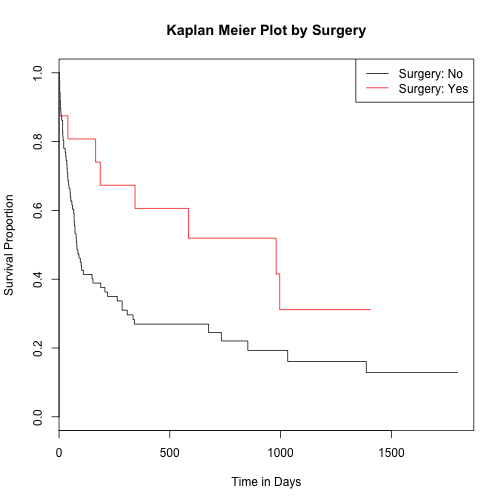
Assess significance of survival by surgery status
survdiff(Surv(time = heart$start, event = heart$event == 1) ~ heart$surgery)
## Call:
## survdiff(formula = Surv(time = heart$start, event = heart$event ==
## 1) ~ heart$surgery)
##
## N Observed Expected (O-E)^2/E (O-E)^2/V
## heart$surgery=0 143 66 58.7 0.902 4.56
## heart$surgery=1 29 9 16.3 3.255 4.56
##
## Chisq= 4.6 on 1 degrees of freedom, p= 0.0328
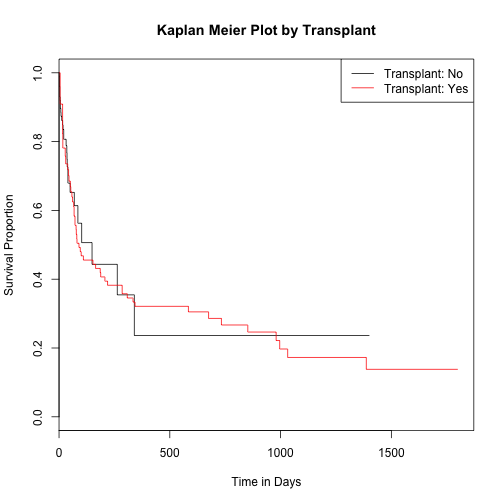
Survival analysis by transplant status
heart$transplant <- as.factor(heart$transplant)
surv_fit3 <- survfit(survival ~ heart$transplant)
plot(surv_fit3, main="Kaplan Meier Plot by Transplant", xlab="Time in Days", ylab="Survival Proportion", col=c(1:2))
legend('topright', c("Transplant: No","Transplant: Yes"), lty=1, col=c(1:2))
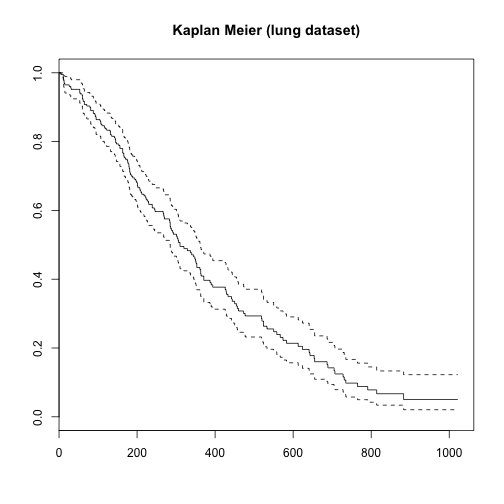
Performing survival analysis with lung dataset
survival_lung <- Surv(time = lung$time, event = lung$status == 2, type='right')
surv_fit3 <- survfit(survival_lung ~ 1)
plot(surv_fit3, main = "Kaplan Meier (lung dataset)")
Survival analysis by sex
lung$sex <- as.factor(lung$sex)
surv_fit4 <- survfit(survival_lung ~ lung$sex)
plot(surv_fit4, main="Kaplan Meier Plot by Sex", xlab="Time in Days", ylab="Survival Proportion", col=c(1:2))
legend('topright', c("Male","Female"), lty=1, col=c(1:2))
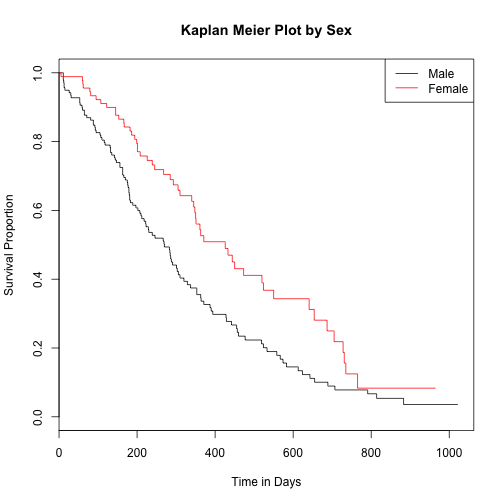
Assess significance of survival by sex
survdiff(Surv(time = lung$time, event = lung$status == 2) ~ lung$sex)
## Call:
## survdiff(formula = Surv(time = lung$time, event = lung$status ==
## 2) ~ lung$sex)
##
## N Observed Expected (O-E)^2/E (O-E)^2/V
## lung$sex=1 138 112 91.6 4.55 10.3
## lung$sex=2 90 53 73.4 5.68 10.3
##
## Chisq= 10.3 on 1 degrees of freedom, p= 0.00131
Survival analysis by ECOG performance score
lung$ph.ecog <- as.factor(lung$ph.ecog) #ECOG performance score (0=good 5=dead)
surv_fit4 <- survfit(survival_lung ~ lung$ph.ecog)
plot(surv_fit4, main="Kaplan Meier Plot by ECOG Performance", xlab="Time in Days", ylab="Survival Proportion", col=c(1:5))
legend('topright', c("Ecog=0","Ecog=1", "Ecog=2", "Ecog=3"), lty=1, col=c(1:5))
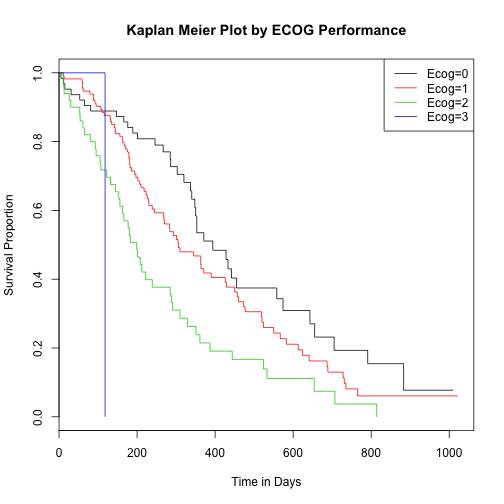
Assess significance of survival by ECOG
survdiff(Surv(time = lung$time, event = lung$status == 2) ~ lung$ph.ecog)
## Call:
## survdiff(formula = Surv(time = lung$time, event = lung$status ==
## 2) ~ lung$ph.ecog)
##
## n=227, 1 observation deleted due to missingness.
##
## N Observed Expected (O-E)^2/E (O-E)^2/V
## lung$ph.ecog=0 63 37 54.153 5.4331 8.2119
## lung$ph.ecog=1 113 82 83.528 0.0279 0.0573
## lung$ph.ecog=2 50 44 26.147 12.1893 14.6491
## lung$ph.ecog=3 1 1 0.172 3.9733 4.0040
##
## Chisq= 22 on 3 degrees of freedom, p= 6.64e-05





